1831: Cell nucleus first described by Robert Brown
1871: DNA of cells first isolated by Frederich Miescher in pus of wounds
1938: Ribosomes, which means "body of ribose" after these organelles were found to contain the sugar ribose, were first described by Albert Claude and were observed in animal cells which had been infected with Rous sarcoma virus. He then ascertained that noninfected cells also contained these particles and first called them microsomes (which in part they were, because ribosomes are bound to microsomes).

1943: Ribosomes in bacterial cells first described by Luria, Delbruck and Anderson.
1950: George Palade and Keith Porter using Electron Microscopy describe Ribosomes when they were at Rockefeller Institute (part of reason for Palade's Nobel prize in 1974), and noticed their location on the endoplasmic reticulum and free in cytoplasm. (ref.)
1953: Watson and Crick describe the double-helix structure of DNA (click here to read their original paper).
2003: Completion of sequencing the human genome.

Some cool info on nuclear pores here

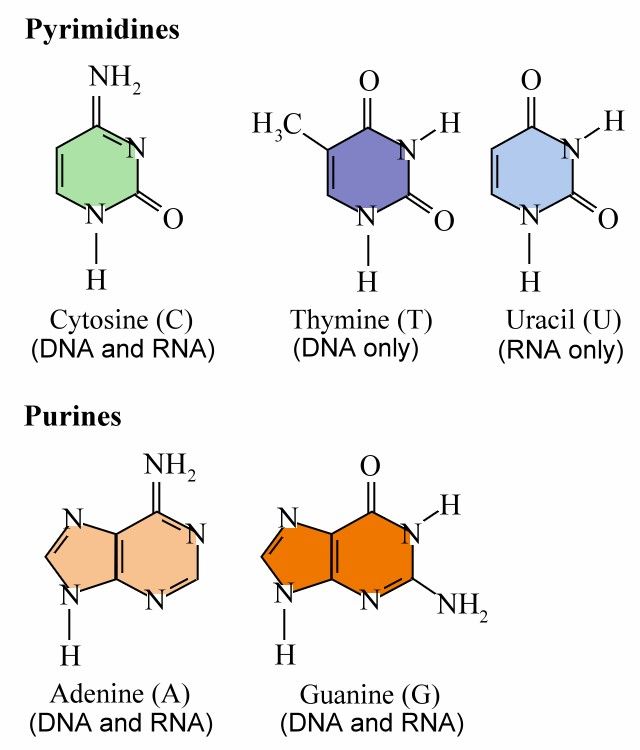
DNA or deoxyribonucleic acid is a large molecule structured from chains of repeating units of the sugar deoxyribose and phosphate linked to four different bases: Adenine, Thymine, Guanine, and Cytosine. Ribonucleic acid (RNA) is a single-stranded copy of DNA, in which the Thymine is replaced by Uricil; (More information on transcription is available here).
During mitosis (cell division), each DNA molecule is wrapped around histones and wrapped further up into chromatin (more info). During interphase (between divisions), chromatin is more extended, a form used for expression genetic information. The DNA is loosely held in the nucleus. (see picture above of cross-sectional view of nuclear envelope). (ref.)
(Click here for a good picture of the bonding between G-C and A-T pairs (ref.)
John Kyrk's website has great Flash animations of DNA's structure, replication, transcription and RNA translation. And From chromosome to protein: animated essential cell function" is one of the best overviews of how everything fits together that I've come across. All are accurately done and illustrate these concepts vividly. Check them out - highly recommended!
As well, this protein synthesis movie is a cool little animation that provides a bit more detail on how protein is synthesized, including the role of enzymes during a step called acylation, where tRNA molecules are linked to their respective amino acids, forming complexes called aminoacyl-tRNAs.
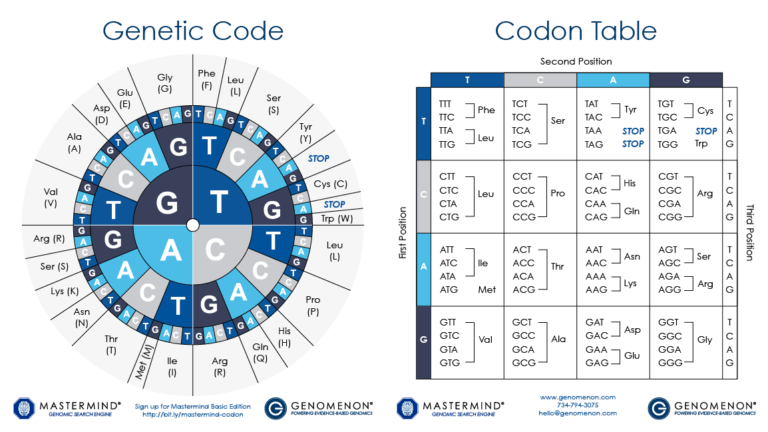 DNA encodes the unique nature of different organisms by specifying the precise structure of each protein in a cell. In analogy to DNA, proteins are made from a linear sequence of amino acids, and the exact sequence of amino acids is what determines the function of the protein. A DNA sequence is translated into the protein sequence by a code, where a triplet of bases (a codon) specifies a single amino acid; some codons specify the end of the protein. As a cell "reads" the DNA instructions, it builds a protein by adding successive amino acids one at a time, as defined by the codons. With 64 possible codons (4 DNA bases in the 3 positions of the codon, or 43) and only 20 standard amino acids, some amino acids can be specified by more than one codon. (ref.)
Some key words:
Transcription = DNA → RNA
Translation = RNA → protein
Polypeptide = a molecule made of a sequence of amino acids linked by peptide bonds. Proteins are made up of one or more polypeptide molecules.
Nucleotide: The basic unit for DNA and RNA is the nucleotides, which consist of three components: the nitrogen bases, the ribose sugars, and the phosphates. ((ref.)
5'[5 prime] and 3'[3 prime]: The 5 and 3 refer to the number of Carbon molecules on the end of a nucleotide. The DNA molecule is made of two asymmetric strands, one end of which terminates with a nucleotide with 5 carbons (the 5' end) and the other which terminates with a nucleotide with 3 carbons. People who work with DNA use the term 5' and 3' to identify a particular end of one of these strands. Within a cell, the enzymes that perform replication and transcription read DNA in the "3' to 5' direction", while the enzymes that perform translation read in the opposite direction (on RNA). (ref.)
Types of RNA (From John Kimball's biology pages)
Several types of RNA are synthesized:
DNA encodes the unique nature of different organisms by specifying the precise structure of each protein in a cell. In analogy to DNA, proteins are made from a linear sequence of amino acids, and the exact sequence of amino acids is what determines the function of the protein. A DNA sequence is translated into the protein sequence by a code, where a triplet of bases (a codon) specifies a single amino acid; some codons specify the end of the protein. As a cell "reads" the DNA instructions, it builds a protein by adding successive amino acids one at a time, as defined by the codons. With 64 possible codons (4 DNA bases in the 3 positions of the codon, or 43) and only 20 standard amino acids, some amino acids can be specified by more than one codon. (ref.)
Some key words:
Transcription = DNA → RNA
Translation = RNA → protein
Polypeptide = a molecule made of a sequence of amino acids linked by peptide bonds. Proteins are made up of one or more polypeptide molecules.
Nucleotide: The basic unit for DNA and RNA is the nucleotides, which consist of three components: the nitrogen bases, the ribose sugars, and the phosphates. ((ref.)
5'[5 prime] and 3'[3 prime]: The 5 and 3 refer to the number of Carbon molecules on the end of a nucleotide. The DNA molecule is made of two asymmetric strands, one end of which terminates with a nucleotide with 5 carbons (the 5' end) and the other which terminates with a nucleotide with 3 carbons. People who work with DNA use the term 5' and 3' to identify a particular end of one of these strands. Within a cell, the enzymes that perform replication and transcription read DNA in the "3' to 5' direction", while the enzymes that perform translation read in the opposite direction (on RNA). (ref.)
Types of RNA (From John Kimball's biology pages)
Several types of RNA are synthesized:
Transfer RNA (tRNA)


(tRNA pictures are from Review of the Universe - very cool website.)
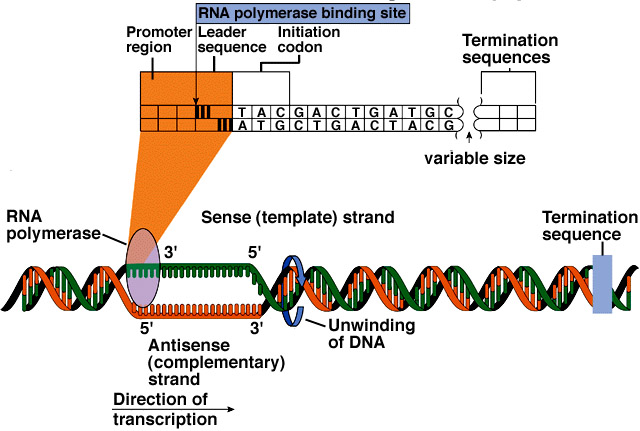 From NBII's Basic Genetics and Cell Biology:
Humans are built from an estimated 20,000-35,000 proteins, yet, only a small percentage of the 3 billion bases in the human genome codes for these proteins. The discrete sections of DNA that encode proteins are referred to as "genes." A gene contains special codons that tell the cell where the gene - and hence, the protein - start and stop. Between these signals, the regions of the DNA that code for amino acids (the exons) are separated by a variable number and length of non-coding DNA sequence (the intervening sequence, or introns).
Protein production is a highly regulated process. For example, a cell does not want to waste energy making the proteins needed for cell division if it is busy with other functions, such as secreting a hormone. This process of turning a gene on and off depending on the cell's need for a particular set of proteins is referred to as regulation of gene expression. Certain proteins, and even other regions of DNA, physically bind to the DNA sequence surrounding a gene to affect its expression. This interaction occurs at regions of the DNA with apt names, such as promoters or enhancers of gene expression. (more on gene transcription...)
Gene regulation is an essential part of life. Since every cell in an organism contains the same genetic blueprint, different cell types are created by turning on different genes at different times during development. In fact, it is differential gene expression that allows stem cells to become unique cell types. Gene regulation is also critical for cellular response to metabolic needs.
There are a number of different ways that are used to control which genes are activated (and thus which proteins, etc. are generated), and the rate at which the resulting molecules that are created at. The most important and widely used mechanism is by altering the rate of transcription of the gene.
Alternative mechanisms include:
From NBII's Basic Genetics and Cell Biology:
Humans are built from an estimated 20,000-35,000 proteins, yet, only a small percentage of the 3 billion bases in the human genome codes for these proteins. The discrete sections of DNA that encode proteins are referred to as "genes." A gene contains special codons that tell the cell where the gene - and hence, the protein - start and stop. Between these signals, the regions of the DNA that code for amino acids (the exons) are separated by a variable number and length of non-coding DNA sequence (the intervening sequence, or introns).
Protein production is a highly regulated process. For example, a cell does not want to waste energy making the proteins needed for cell division if it is busy with other functions, such as secreting a hormone. This process of turning a gene on and off depending on the cell's need for a particular set of proteins is referred to as regulation of gene expression. Certain proteins, and even other regions of DNA, physically bind to the DNA sequence surrounding a gene to affect its expression. This interaction occurs at regions of the DNA with apt names, such as promoters or enhancers of gene expression. (more on gene transcription...)
Gene regulation is an essential part of life. Since every cell in an organism contains the same genetic blueprint, different cell types are created by turning on different genes at different times during development. In fact, it is differential gene expression that allows stem cells to become unique cell types. Gene regulation is also critical for cellular response to metabolic needs.
There are a number of different ways that are used to control which genes are activated (and thus which proteins, etc. are generated), and the rate at which the resulting molecules that are created at. The most important and widely used mechanism is by altering the rate of transcription of the gene.
Alternative mechanisms include:
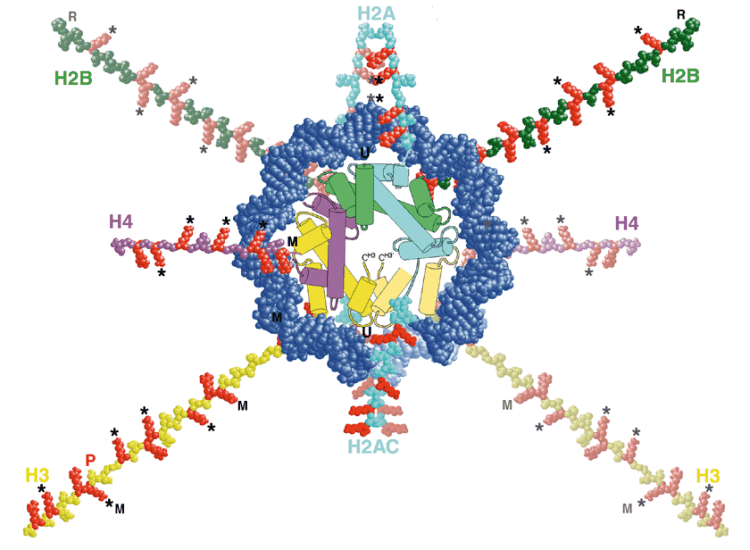 The preceding blog entry touched on how the DNA molecule is wound around histones, then further wound into a fibre of chromatin which is then finally wound up to form chromosomes. The way DNA winds around histones is something that is undergoing active research, because it looks like it may play an important role in regulating transcription.
Scientists believe that histone modifications are crucial actors in the activation and repression of gene expression. Histones help to package DNA, the hereditary material of life, into each cell's nucleus. The double-helical strand of DNA wraps around a ball of histones consisting of four distinct proteins: H2A, H2B, H3 and H4. [The genes that encode the H3 and H4 proteins used in histones are so fundamental to the actual storage of the genome that they have a zero effective mutation rate. Almost the exact same genes are found in humans, chickens, grass, and mould. (ref.)]
The preceding blog entry touched on how the DNA molecule is wound around histones, then further wound into a fibre of chromatin which is then finally wound up to form chromosomes. The way DNA winds around histones is something that is undergoing active research, because it looks like it may play an important role in regulating transcription.
Scientists believe that histone modifications are crucial actors in the activation and repression of gene expression. Histones help to package DNA, the hereditary material of life, into each cell's nucleus. The double-helical strand of DNA wraps around a ball of histones consisting of four distinct proteins: H2A, H2B, H3 and H4. [The genes that encode the H3 and H4 proteins used in histones are so fundamental to the actual storage of the genome that they have a zero effective mutation rate. Almost the exact same genes are found in humans, chickens, grass, and mould. (ref.)]
 In 1997, Roeder and Wei Gu, then a postdoc in Roeder's lab, showed that an enzyme called p300/CBP, known to modify the chemical composition of histone tails, serves as a transcriptional coactivator for p53. Coactivators are regulatory proteins that, together with activator proteins, are required to turn genes on.
...
In 2002, An, Roeder and their Rockefeller colleagues provided important new information about the role of histone tails in gene activation. Scientists knew that histone tails repressed gene activation by preventing transcription factors -- proteins that help read out the information encoded in DNA -- from gaining access to DNA. An and colleagues, using a test tube system of coiled up chromatin created from engineered or recombinant histones and DNA, showed that -- as Allfrey and Allis had predicted -- histone tails and associated modifications are required for reversing the repression of transcription and that p300 plays an important role in this process as a histone-modifying enzyme. (ref.)
Until a few years ago was it generally assumed that the histones take part in the selective transcription of single DNA segments (genes or groups of genes). This proved to be wrong: a group of acidic nuclear proteins (non-histones) fulfils this function. Non-histones is a collective term for a number of regulators that are acidic when compared to the strongly alkaline histones. Otherwise is their amino acid composition within the usual framework.(ref.)
The way DNA loops around the histones is controlled by the characteristics of the histones involved, as shown below:
In 1997, Roeder and Wei Gu, then a postdoc in Roeder's lab, showed that an enzyme called p300/CBP, known to modify the chemical composition of histone tails, serves as a transcriptional coactivator for p53. Coactivators are regulatory proteins that, together with activator proteins, are required to turn genes on.
...
In 2002, An, Roeder and their Rockefeller colleagues provided important new information about the role of histone tails in gene activation. Scientists knew that histone tails repressed gene activation by preventing transcription factors -- proteins that help read out the information encoded in DNA -- from gaining access to DNA. An and colleagues, using a test tube system of coiled up chromatin created from engineered or recombinant histones and DNA, showed that -- as Allfrey and Allis had predicted -- histone tails and associated modifications are required for reversing the repression of transcription and that p300 plays an important role in this process as a histone-modifying enzyme. (ref.)
Until a few years ago was it generally assumed that the histones take part in the selective transcription of single DNA segments (genes or groups of genes). This proved to be wrong: a group of acidic nuclear proteins (non-histones) fulfils this function. Non-histones is a collective term for a number of regulators that are acidic when compared to the strongly alkaline histones. Otherwise is their amino acid composition within the usual framework.(ref.)
The way DNA loops around the histones is controlled by the characteristics of the histones involved, as shown below:
 More...
More...Role of the Ribosome: The route from the DNA code to the protein
Visualizing Cell Biology
Nucleic Acid Nomenclature and Structure
Computational Biology:
Bhageerath: an energy based web enabled computer software suite for limiting the search space of tertiary structures of small globular proteins
Molecular modeling of the chromatosome particle
Transcription Regulation in Eukaryotes
Linking structure to function with histone H1
The Endoplasmic Reticulum as a Protein-Folding Compartment: excerpt: Many human diseases, such as cystic fibrosis and Alzheimer's disease, result from improper protein folding. Our studies into the fundamental processes of how the Endoplasmic Reticulum coordinates protein folding and its response to unfolded proteins will provide new avenues to treat the diseases of protein misfolding. Our recent studies also demonstrate, however, that the unfolded protein response signaling pathways are also essential for cell differentiation and cell function under normal physiological conditions.
Special thanks to DNA 2.0 and Mrs. King's Bio Web for the use of the Codon Table wheel image.
No comments:
Post a Comment