Nanobiology Notes
You can see, if you look closely, forever...
Sunday, February 19, 2006
Getting started
Amazing things happen at the boundary between quantum physics and the 3D world of Netwonian physics. Like life, for example. I've been reading a lot about recent research in molecular biology and the basic building blocks of life at the molecular nanoscopic level. The following are the notes I've been making along the way to consolidate my understanding, including quotes and pictures from a lot of very cool web sites and links to multimedia presentations that vividly show how amazing the world is at the nanometer scale. Hope you find them interesting, fun and thought provoking.
Just add water...
Membranes - Keeping the inside in and the outside out
From Stuart Kauffman's essay "What is Life?" in the book "The next fifty years": An enclosed gas in a thermodynamically isolated box can do no work. But if the box is divided into two parts by a membrane, then one part can do work on the other part; for example, if the gas pressure is higher in the first part, the membrane will bulge into the second part, doing mechanical work on it. Thus work cannot be achieved in the universe unless the universe is divided into at least two regions. Furthermore, just where did the membrane come from?
From Henry Jakubowski's page at St. John's University: How would you orient this molecule in water? There are several possible ways. A small number of these molecules might be soluble in water as above. Mostly, however, the nonpolar tails wants to get out of the water, while the polar head like to stay in the water. Again, some of the molecules migrate to the surface of the water, with the nonpolar tails sticking out into air, away from water, to form a monolayer on the top of the water. Others will self- aggregate, through Inter-Molecular Forces (IMF's) to form a bilayer or membrane. Because there are two tails per head group, the tails can't pack together as tightly. Imagine the bilayer or membrane curving around and eventually meeting. A structure like this would look like a small biological cell. ... The interior of this little cell, or liposome, is filled with water.
Now, cell membranes are not rigid or impermeable. In fact, things like proteins with one hydrophobic end can push into them and get embedded in them fairly easily. Ions (like Na+ or K+, for example) are not able to pass through the membrane. Also, the acidity of the water inside the liposome can become different than outside of the liposome. This sets up the conditions for molecules to diffuse through the membrane, as ions are driven by the concentration gradient to spread to regions of lower concentration. i.e. it sets up the conditions needed to get some work done!
This still leaves Stuart Kauffman's question - "Just where did the membrane come from?"
From First Cell by Carl Zimmer: Life is chemical interaction, and for that interaction to occur, life’s molecules must be close to one another. Without a physical boundary of some sort, without a skin, a bark, or a cell membrane, an organism is nothing more than a diffusing blur of molecules. To explain how the first creature came to be, you have to explain how its innards got to be distinguished from its surroundings. In other words, you’ve got to explain how the first single- celled creature got encapsulated in a cell.
...
A cell membrane’s importance to life is often underappreciated, says David Deamer (University of California at Santa Cruz). People say, ‘Well, it’s just a little bag.’ But it’s much more. It’s the interface between life and everything that’s outside. The membrane of any cell has to do many things at once. It has to be impermeable enough to keep essential things (like DNA) in and harmful things (like viruses and poisons) out. Yet a cell membrane can’t form a perfect seal. It has to be able to flush out waste and heat from its own system and take in nutrients from the surrounding medium. And the first cell membrane, like the membranes of many single-celled organisms today, probably had to be able to collect energy as well. No sense repeating the whole article here - it's a fascinating read on how life might have gotten started - and membranes look like they were the key.
Fun with Molecular Origami
When you start reading about how the brain works and how cells work, you come across a number of key words over and over. Words like 'ion channel', 'neurotransmitter', 'receptor'. Well, what the heck are these things, really? Finding the answer to that question has dragged me through a heck of a lot of interesting territory. The short answer is 'protein'. They are all made of protein. So, before diving into receptors and stuff, here's some info I've found on proteins:
Biochemistry toolkit
Background info:
Review of Chemistry,
The Chemistry of Life,
Macromolecules: Sugars and Lipids,
Macromolecules: Proteins and Nucleic Acids
(Thank you to Frank Orme for writing and posting these excellent, readable, concise notes!)
From all of that, we know that proteins are built out of a 'toolkit' of 20 amino acids. An amino acid looks like this:

i.e. a central carbon attached to a carboxylate (the COOH), an amino (the NH2) and a hydrogen atom. The differences between the 20 types are in the side groups that can be designated as R. R can either be hydrophobic or hydrophilic.
 (Note: I've recently updated this image to better align with published results such as Pommié, C. et al., J. Mol. Recognit., 17, 17-32 (2004))
Protein is made by stringing together a bunch of these amino acids with peptide bonds.
(Note: I've recently updated this image to better align with published results such as Pommié, C. et al., J. Mol. Recognit., 17, 17-32 (2004))
Protein is made by stringing together a bunch of these amino acids with peptide bonds.

(Ref: Georgia State University's HyperPhysics site) The body builds around 25000 different types of proteins, some of which act as nano-scale engines (e.g. in muscles), others containing 500-1000 amino acids act as enzymes, some are used as messengers within the cell or between cells, and still others that have both a hydrophobic amino acid side group and a hydrophilic amino acid side group embed themselves in the cell wall and form either ion channels or receptors. (Hey, we're finally getting to the point...) The 3D shape of the protein controls how other molecules interact with the protein. Check out this great flash animation that shows all of the amino acids, the formation of peptide bonds, the formation of alpha helixes and beta sheets and visualization of what it means to 'fold' a protein. Folding of proteins can be examined using 3D Electron Microscopy. "Folds are not "fixed" structures but rather flexible definitions related with the number of secondary structure elements and its spatial distribution." The CATH protein structure classification database catalogs proteins by Class, Architecture, Topology (fold family) and Homologous Superfamily (proteins thought to share a common ancestor).
 Unraveling the Mystery of Protein Folding provides a fascinating overview of recent discoveries about the 3D mechanics of proteins, including the role that improperly folded proteins have been found to play in Alzheimer's disease. IBM's Blue Gene supercomputer is one of the most powerful tools being used in this area of study. It allows researchers to model the 3D forces at play that result in protein folding.
Examples:
Unraveling the Mystery of Protein Folding provides a fascinating overview of recent discoveries about the 3D mechanics of proteins, including the role that improperly folded proteins have been found to play in Alzheimer's disease. IBM's Blue Gene supercomputer is one of the most powerful tools being used in this area of study. It allows researchers to model the 3D forces at play that result in protein folding.
Examples:
G Protein-Coupled Receptors (GPCR) in a membrane environment
Lipids critical to cell division and fusion
More... From the excellent Medical Biochemistry page at Indian University: Protein Structure - covers hydrogen bonding and other forces involved in folding proteins. Charge Densities, Hydrogen Bonding and Drug Design shows electron density contributions from lone pair, covalent and hydrogen bond electrons.
i.e. a central carbon attached to a carboxylate (the COOH), an amino (the NH2) and a hydrogen atom. The differences between the 20 types are in the side groups that can be designated as R. R can either be hydrophobic or hydrophilic.
(Ref: Georgia State University's HyperPhysics site) The body builds around 25000 different types of proteins, some of which act as nano-scale engines (e.g. in muscles), others containing 500-1000 amino acids act as enzymes, some are used as messengers within the cell or between cells, and still others that have both a hydrophobic amino acid side group and a hydrophilic amino acid side group embed themselves in the cell wall and form either ion channels or receptors. (Hey, we're finally getting to the point...) The 3D shape of the protein controls how other molecules interact with the protein. Check out this great flash animation that shows all of the amino acids, the formation of peptide bonds, the formation of alpha helixes and beta sheets and visualization of what it means to 'fold' a protein. Folding of proteins can be examined using 3D Electron Microscopy. "Folds are not "fixed" structures but rather flexible definitions related with the number of secondary structure elements and its spatial distribution." The CATH protein structure classification database catalogs proteins by Class, Architecture, Topology (fold family) and Homologous Superfamily (proteins thought to share a common ancestor).
G Protein-Coupled Receptors (GPCR) in a membrane environment
Lipids critical to cell division and fusion
More... From the excellent Medical Biochemistry page at Indian University: Protein Structure - covers hydrogen bonding and other forces involved in folding proteins. Charge Densities, Hydrogen Bonding and Drug Design shows electron density contributions from lone pair, covalent and hydrogen bond electrons.
Protein formation: Codones, Histones and Ribosomes
We've come a long way...
1831: Cell nucleus first described by Robert Brown
1871: DNA of cells first isolated by Frederich Miescher in pus of wounds
1938: Ribosomes, which means "body of ribose" after these organelles were found to contain the sugar ribose, were first described by Albert Claude and were observed in animal cells which had been infected with Rous sarcoma virus. He then ascertained that noninfected cells also contained these particles and first called them microsomes (which in part they were, because ribosomes are bound to microsomes).
1943: Ribosomes in bacterial cells first described by Luria, Delbruck and Anderson.
1950: George Palade and Keith Porter using Electron Microscopy describe Ribosomes when they were at Rockefeller Institute (part of reason for Palade's Nobel prize in 1974), and noticed their location on the endoplasmic reticulum and free in cytoplasm. (ref.)
1953: Watson and Crick describe the double-helix structure of DNA (click here to read their original paper).
2003: Completion of sequencing the human genome.

Some cool info on nuclear pores here

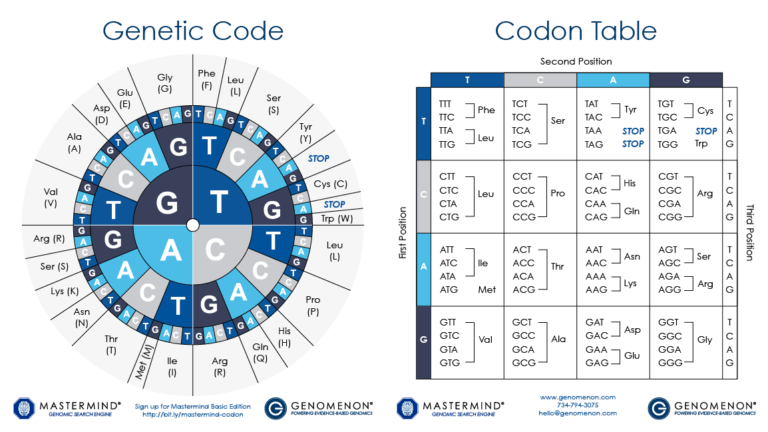 DNA encodes the unique nature of different organisms by specifying the precise structure of each protein in a cell. In analogy to DNA, proteins are made from a linear sequence of amino acids, and the exact sequence of amino acids is what determines the function of the protein. A DNA sequence is translated into the protein sequence by a code, where a triplet of bases (a codon) specifies a single amino acid; some codons specify the end of the protein. As a cell "reads" the DNA instructions, it builds a protein by adding successive amino acids one at a time, as defined by the codons. With 64 possible codons (4 DNA bases in the 3 positions of the codon, or 43) and only 20 standard amino acids, some amino acids can be specified by more than one codon. (ref.)
Some key words:
Transcription = DNA → RNA
Translation = RNA → protein
Polypeptide = a molecule made of a sequence of amino acids linked by peptide bonds. Proteins are made up of one or more polypeptide molecules.
Nucleotide: The basic unit for DNA and RNA is the nucleotides, which consist of three components: the nitrogen bases, the ribose sugars, and the phosphates. ((ref.)
5'[5 prime] and 3'[3 prime]: The 5 and 3 refer to the number of Carbon molecules on the end of a nucleotide. The DNA molecule is made of two asymmetric strands, one end of which terminates with a nucleotide with 5 carbons (the 5' end) and the other which terminates with a nucleotide with 3 carbons. People who work with DNA use the term 5' and 3' to identify a particular end of one of these strands. Within a cell, the enzymes that perform replication and transcription read DNA in the "3' to 5' direction", while the enzymes that perform translation read in the opposite direction (on RNA). (ref.)
Types of RNA (From John Kimball's biology pages)
Several types of RNA are synthesized:
DNA encodes the unique nature of different organisms by specifying the precise structure of each protein in a cell. In analogy to DNA, proteins are made from a linear sequence of amino acids, and the exact sequence of amino acids is what determines the function of the protein. A DNA sequence is translated into the protein sequence by a code, where a triplet of bases (a codon) specifies a single amino acid; some codons specify the end of the protein. As a cell "reads" the DNA instructions, it builds a protein by adding successive amino acids one at a time, as defined by the codons. With 64 possible codons (4 DNA bases in the 3 positions of the codon, or 43) and only 20 standard amino acids, some amino acids can be specified by more than one codon. (ref.)
Some key words:
Transcription = DNA → RNA
Translation = RNA → protein
Polypeptide = a molecule made of a sequence of amino acids linked by peptide bonds. Proteins are made up of one or more polypeptide molecules.
Nucleotide: The basic unit for DNA and RNA is the nucleotides, which consist of three components: the nitrogen bases, the ribose sugars, and the phosphates. ((ref.)
5'[5 prime] and 3'[3 prime]: The 5 and 3 refer to the number of Carbon molecules on the end of a nucleotide. The DNA molecule is made of two asymmetric strands, one end of which terminates with a nucleotide with 5 carbons (the 5' end) and the other which terminates with a nucleotide with 3 carbons. People who work with DNA use the term 5' and 3' to identify a particular end of one of these strands. Within a cell, the enzymes that perform replication and transcription read DNA in the "3' to 5' direction", while the enzymes that perform translation read in the opposite direction (on RNA). (ref.)
Types of RNA (From John Kimball's biology pages)
Several types of RNA are synthesized:
 From NBII's Basic Genetics and Cell Biology:
Humans are built from an estimated 20,000-35,000 proteins, yet, only a small percentage of the 3 billion bases in the human genome codes for these proteins. The discrete sections of DNA that encode proteins are referred to as "genes." A gene contains special codons that tell the cell where the gene - and hence, the protein - start and stop. Between these signals, the regions of the DNA that code for amino acids (the exons) are separated by a variable number and length of non-coding DNA sequence (the intervening sequence, or introns).
Protein production is a highly regulated process. For example, a cell does not want to waste energy making the proteins needed for cell division if it is busy with other functions, such as secreting a hormone. This process of turning a gene on and off depending on the cell's need for a particular set of proteins is referred to as regulation of gene expression. Certain proteins, and even other regions of DNA, physically bind to the DNA sequence surrounding a gene to affect its expression. This interaction occurs at regions of the DNA with apt names, such as promoters or enhancers of gene expression. (more on gene transcription...)
Gene regulation is an essential part of life. Since every cell in an organism contains the same genetic blueprint, different cell types are created by turning on different genes at different times during development. In fact, it is differential gene expression that allows stem cells to become unique cell types. Gene regulation is also critical for cellular response to metabolic needs.
There are a number of different ways that are used to control which genes are activated (and thus which proteins, etc. are generated), and the rate at which the resulting molecules that are created at. The most important and widely used mechanism is by altering the rate of transcription of the gene.
Alternative mechanisms include:
From NBII's Basic Genetics and Cell Biology:
Humans are built from an estimated 20,000-35,000 proteins, yet, only a small percentage of the 3 billion bases in the human genome codes for these proteins. The discrete sections of DNA that encode proteins are referred to as "genes." A gene contains special codons that tell the cell where the gene - and hence, the protein - start and stop. Between these signals, the regions of the DNA that code for amino acids (the exons) are separated by a variable number and length of non-coding DNA sequence (the intervening sequence, or introns).
Protein production is a highly regulated process. For example, a cell does not want to waste energy making the proteins needed for cell division if it is busy with other functions, such as secreting a hormone. This process of turning a gene on and off depending on the cell's need for a particular set of proteins is referred to as regulation of gene expression. Certain proteins, and even other regions of DNA, physically bind to the DNA sequence surrounding a gene to affect its expression. This interaction occurs at regions of the DNA with apt names, such as promoters or enhancers of gene expression. (more on gene transcription...)
Gene regulation is an essential part of life. Since every cell in an organism contains the same genetic blueprint, different cell types are created by turning on different genes at different times during development. In fact, it is differential gene expression that allows stem cells to become unique cell types. Gene regulation is also critical for cellular response to metabolic needs.
There are a number of different ways that are used to control which genes are activated (and thus which proteins, etc. are generated), and the rate at which the resulting molecules that are created at. The most important and widely used mechanism is by altering the rate of transcription of the gene.
Alternative mechanisms include:
 The preceding blog entry touched on how the DNA molecule is wound around histones, then further wound into a fibre of chromatin which is then finally wound up to form chromosomes. The way DNA winds around histones is something that is undergoing active research, because it looks like it may play an important role in regulating transcription.
Scientists believe that histone modifications are crucial actors in the activation and repression of gene expression. Histones help to package DNA, the hereditary material of life, into each cell's nucleus. The double-helical strand of DNA wraps around a ball of histones consisting of four distinct proteins: H2A, H2B, H3 and H4. [The genes that encode the H3 and H4 proteins used in histones are so fundamental to the actual storage of the genome that they have a zero effective mutation rate. Almost the exact same genes are found in humans, chickens, grass, and mould. (ref.)]
The preceding blog entry touched on how the DNA molecule is wound around histones, then further wound into a fibre of chromatin which is then finally wound up to form chromosomes. The way DNA winds around histones is something that is undergoing active research, because it looks like it may play an important role in regulating transcription.
Scientists believe that histone modifications are crucial actors in the activation and repression of gene expression. Histones help to package DNA, the hereditary material of life, into each cell's nucleus. The double-helical strand of DNA wraps around a ball of histones consisting of four distinct proteins: H2A, H2B, H3 and H4. [The genes that encode the H3 and H4 proteins used in histones are so fundamental to the actual storage of the genome that they have a zero effective mutation rate. Almost the exact same genes are found in humans, chickens, grass, and mould. (ref.)]
 In 1997, Roeder and Wei Gu, then a postdoc in Roeder's lab, showed that an enzyme called p300/CBP, known to modify the chemical composition of histone tails, serves as a transcriptional coactivator for p53. Coactivators are regulatory proteins that, together with activator proteins, are required to turn genes on.
...
In 2002, An, Roeder and their Rockefeller colleagues provided important new information about the role of histone tails in gene activation. Scientists knew that histone tails repressed gene activation by preventing transcription factors -- proteins that help read out the information encoded in DNA -- from gaining access to DNA. An and colleagues, using a test tube system of coiled up chromatin created from engineered or recombinant histones and DNA, showed that -- as Allfrey and Allis had predicted -- histone tails and associated modifications are required for reversing the repression of transcription and that p300 plays an important role in this process as a histone-modifying enzyme. (ref.)
Until a few years ago was it generally assumed that the histones take part in the selective transcription of single DNA segments (genes or groups of genes). This proved to be wrong: a group of acidic nuclear proteins (non-histones) fulfils this function. Non-histones is a collective term for a number of regulators that are acidic when compared to the strongly alkaline histones. Otherwise is their amino acid composition within the usual framework.(ref.)
The way DNA loops around the histones is controlled by the characteristics of the histones involved, as shown below:
In 1997, Roeder and Wei Gu, then a postdoc in Roeder's lab, showed that an enzyme called p300/CBP, known to modify the chemical composition of histone tails, serves as a transcriptional coactivator for p53. Coactivators are regulatory proteins that, together with activator proteins, are required to turn genes on.
...
In 2002, An, Roeder and their Rockefeller colleagues provided important new information about the role of histone tails in gene activation. Scientists knew that histone tails repressed gene activation by preventing transcription factors -- proteins that help read out the information encoded in DNA -- from gaining access to DNA. An and colleagues, using a test tube system of coiled up chromatin created from engineered or recombinant histones and DNA, showed that -- as Allfrey and Allis had predicted -- histone tails and associated modifications are required for reversing the repression of transcription and that p300 plays an important role in this process as a histone-modifying enzyme. (ref.)
Until a few years ago was it generally assumed that the histones take part in the selective transcription of single DNA segments (genes or groups of genes). This proved to be wrong: a group of acidic nuclear proteins (non-histones) fulfils this function. Non-histones is a collective term for a number of regulators that are acidic when compared to the strongly alkaline histones. Otherwise is their amino acid composition within the usual framework.(ref.)
The way DNA loops around the histones is controlled by the characteristics of the histones involved, as shown below:
 More...
More...
Role of the Ribosome: The route from the DNA code to the protein
Visualizing Cell Biology
Nucleic Acid Nomenclature and Structure
Computational Biology:
Bhageerath: an energy based web enabled computer software suite for limiting the search space of tertiary structures of small globular proteins
Molecular modeling of the chromatosome particle
Transcription Regulation in Eukaryotes
Linking structure to function with histone H1
The Endoplasmic Reticulum as a Protein-Folding Compartment: excerpt: Many human diseases, such as cystic fibrosis and Alzheimer's disease, result from improper protein folding. Our studies into the fundamental processes of how the Endoplasmic Reticulum coordinates protein folding and its response to unfolded proteins will provide new avenues to treat the diseases of protein misfolding. Our recent studies also demonstrate, however, that the unfolded protein response signaling pathways are also essential for cell differentiation and cell function under normal physiological conditions.
Special thanks to DNA 2.0 and Mrs. King's Bio Web for the use of the Codon Table wheel image.
1831: Cell nucleus first described by Robert Brown
1871: DNA of cells first isolated by Frederich Miescher in pus of wounds
1938: Ribosomes, which means "body of ribose" after these organelles were found to contain the sugar ribose, were first described by Albert Claude and were observed in animal cells which had been infected with Rous sarcoma virus. He then ascertained that noninfected cells also contained these particles and first called them microsomes (which in part they were, because ribosomes are bound to microsomes).
1943: Ribosomes in bacterial cells first described by Luria, Delbruck and Anderson.
1950: George Palade and Keith Porter using Electron Microscopy describe Ribosomes when they were at Rockefeller Institute (part of reason for Palade's Nobel prize in 1974), and noticed their location on the endoplasmic reticulum and free in cytoplasm. (ref.)
1953: Watson and Crick describe the double-helix structure of DNA (click here to read their original paper).
2003: Completion of sequencing the human genome.
Some cool info on nuclear pores here
DNA or deoxyribonucleic acid is a large molecule structured from chains of repeating units of the sugar deoxyribose and phosphate linked to four different bases: Adenine, Thymine, Guanine, and Cytosine. Ribonucleic acid (RNA) is a single-stranded copy of DNA, in which the Thymine is replaced by Uricil; (More information on transcription is available here).
During mitosis (cell division), each DNA molecule is wrapped around histones and wrapped further up into chromatin (more info). During interphase (between divisions), chromatin is more extended, a form used for expression genetic information. The DNA is loosely held in the nucleus. (see picture above of cross-sectional view of nuclear envelope). (ref.)
(Click here for a good picture of the bonding between G-C and A-T pairs (ref.)
John Kyrk's website has great Flash animations of DNA's structure, replication, transcription and RNA translation. And From chromosome to protein: animated essential cell function" is one of the best overviews of how everything fits together that I've come across. All are accurately done and illustrate these concepts vividly. Check them out - highly recommended!
As well, this protein synthesis movie is a cool little animation that provides a bit more detail on how protein is synthesized, including the role of enzymes during a step called acylation, where tRNA molecules are linked to their respective amino acids, forming complexes called aminoacyl-tRNAs.
 DNA encodes the unique nature of different organisms by specifying the precise structure of each protein in a cell. In analogy to DNA, proteins are made from a linear sequence of amino acids, and the exact sequence of amino acids is what determines the function of the protein. A DNA sequence is translated into the protein sequence by a code, where a triplet of bases (a codon) specifies a single amino acid; some codons specify the end of the protein. As a cell "reads" the DNA instructions, it builds a protein by adding successive amino acids one at a time, as defined by the codons. With 64 possible codons (4 DNA bases in the 3 positions of the codon, or 43) and only 20 standard amino acids, some amino acids can be specified by more than one codon. (ref.)
Some key words:
Transcription = DNA → RNA
Translation = RNA → protein
Polypeptide = a molecule made of a sequence of amino acids linked by peptide bonds. Proteins are made up of one or more polypeptide molecules.
Nucleotide: The basic unit for DNA and RNA is the nucleotides, which consist of three components: the nitrogen bases, the ribose sugars, and the phosphates. ((ref.)
5'[5 prime] and 3'[3 prime]: The 5 and 3 refer to the number of Carbon molecules on the end of a nucleotide. The DNA molecule is made of two asymmetric strands, one end of which terminates with a nucleotide with 5 carbons (the 5' end) and the other which terminates with a nucleotide with 3 carbons. People who work with DNA use the term 5' and 3' to identify a particular end of one of these strands. Within a cell, the enzymes that perform replication and transcription read DNA in the "3' to 5' direction", while the enzymes that perform translation read in the opposite direction (on RNA). (ref.)
Types of RNA (From John Kimball's biology pages)
Several types of RNA are synthesized:
DNA encodes the unique nature of different organisms by specifying the precise structure of each protein in a cell. In analogy to DNA, proteins are made from a linear sequence of amino acids, and the exact sequence of amino acids is what determines the function of the protein. A DNA sequence is translated into the protein sequence by a code, where a triplet of bases (a codon) specifies a single amino acid; some codons specify the end of the protein. As a cell "reads" the DNA instructions, it builds a protein by adding successive amino acids one at a time, as defined by the codons. With 64 possible codons (4 DNA bases in the 3 positions of the codon, or 43) and only 20 standard amino acids, some amino acids can be specified by more than one codon. (ref.)
Some key words:
Transcription = DNA → RNA
Translation = RNA → protein
Polypeptide = a molecule made of a sequence of amino acids linked by peptide bonds. Proteins are made up of one or more polypeptide molecules.
Nucleotide: The basic unit for DNA and RNA is the nucleotides, which consist of three components: the nitrogen bases, the ribose sugars, and the phosphates. ((ref.)
5'[5 prime] and 3'[3 prime]: The 5 and 3 refer to the number of Carbon molecules on the end of a nucleotide. The DNA molecule is made of two asymmetric strands, one end of which terminates with a nucleotide with 5 carbons (the 5' end) and the other which terminates with a nucleotide with 3 carbons. People who work with DNA use the term 5' and 3' to identify a particular end of one of these strands. Within a cell, the enzymes that perform replication and transcription read DNA in the "3' to 5' direction", while the enzymes that perform translation read in the opposite direction (on RNA). (ref.)
Types of RNA (From John Kimball's biology pages)
Several types of RNA are synthesized:
Transfer RNA (tRNA)
(tRNA pictures are from Review of the Universe - very cool website.)
Role of the Ribosome: The route from the DNA code to the protein
Visualizing Cell Biology
Nucleic Acid Nomenclature and Structure
Computational Biology:
Bhageerath: an energy based web enabled computer software suite for limiting the search space of tertiary structures of small globular proteins
Molecular modeling of the chromatosome particle
Transcription Regulation in Eukaryotes
Linking structure to function with histone H1
The Endoplasmic Reticulum as a Protein-Folding Compartment: excerpt: Many human diseases, such as cystic fibrosis and Alzheimer's disease, result from improper protein folding. Our studies into the fundamental processes of how the Endoplasmic Reticulum coordinates protein folding and its response to unfolded proteins will provide new avenues to treat the diseases of protein misfolding. Our recent studies also demonstrate, however, that the unfolded protein response signaling pathways are also essential for cell differentiation and cell function under normal physiological conditions.
Special thanks to DNA 2.0 and Mrs. King's Bio Web for the use of the Codon Table wheel image.
Chromosomes: Good things come in very small packages
Cohesin sites (red ovals) are concentrated at the centromere/pericentric region (where the two chromatids are “pinched”), but also occur along the arms of the chromatids. MIT recently discovered that a protein called MEI-S332 regulates the chromosome cohesion, releasing the chromosomes from each other by adding a phosphate to the binding point. These findings are particularly significant given that researchers have found that levels of MEI-S332 are higher than normal in 90% of all breast cancers. According to Clarke, this might mean that when there’s too much of the protein, the chromosomes don’t separate properly, or it might mean that the MEI-S332 gene is mutated on the chromosomes. (ref.) During cell division (mitosis), the two sister chromatids of each of the 46 chromosomes are pulled apart. Protein filaments called microtubules attach the centromeres of the sister chromatids of the chromosomes to the cytoskeleton on opposite sides of the cell. The cytoskeleton is located along the inside of the cell membrane and acts like a nano-scale motor, pulling on the microtubules to line up the chromosomes in the middle of the cell and then literally yanking the sister chromatids apart. There's a great animation by HybridMedicalAnimation of this available here. There's also a fascinating movie of a plant cell undergoing mitosis (more here). It took me a while to appreciate that the structural packaging of DNA differs in bacteria, plants and animals. More:
'From a single double helix to paired double helices and back' The detailed mechanics involved in aligning the chromosomes down the middle of the cell is covered in this article.
Ion Channels: gates in the cell wall
OUTSIDE THE ION FILTER (upper fig.)
Outside the cell membrane the ions are bound to water molecules with certain distances to the oxygen atoms of the water. INSIDE THE ION FILTER (lower fig.)
For the potassium ions the distance to the oxygen atoms in the ion filter is the same as in water. The ion channel is able to strip the potassium ion of its water and allow it to pass at no cost in energy. The sodium ions, which are smaller, do not fit in between the oxygen atoms in the filter. This prevents them from entering the channel.
From NIH - Molecular Biology of the Cell: Many carrier proteins allow solutes to cross the membrane only passively ("downhill"), a process called passive transport, or facilitated diffusion. In the case of transport of a single uncharged molecule, it is simply the difference in its concentration on the two sides of the membrane (its concentration gradient) that drives passive transport and determines its direction. Ionophores are small hydrophobic molecules that dissolve in lipid bilayers and increase their permeability to specific inorganic ions. ... [They] operate by shielding the charge of the transported ion so that it can penetrate the hydrophobic interior of the lipid bilayer. Since ionophores are not coupled to energy sources, they permit the net movement of ions only down their electrochemical gradients. Valinomycin is an example of a mobile ion carrier. It is a ring-shaped polymer that transports K+ down its electrochemical gradient by picking up K+ on one side of the membrane, diffusing across the bilayer, and releasing K+ on the other side.
Similarly, FCCP, a mobile ion carrier that makes membranes selectively leaky to H+, is often used to dissipate the H+ electrochemical gradient across the mitochondrial inner membrane, thereby blocking mitochondrial ATP production. A23187 is yet another example of a mobile ion carrier, only it transports divalent cations such as Ca2+ and Mg2+. When cells are exposed to A23187, Ca2+ enters the cytosol from the extracellular fluid down a steep electrochemical gradient. Accordingly, this ionophore is widely used to increase the concentration of free Ca2+ in the cytosol, thereby mimicking certain cell-signaling mechanisms. (more...) Ion Pumps
Life and Ligands
One of the things I keep coming across is the expression 'ligand', as in 'ligand gated ion channel' or 'receptor ligand'. So I did some digging. The dictionary says it's "An ion, a molecule, or a molecular group that binds to another chemical entity to form a larger complex." But I've started to appreciate that it's a lot more than that. From what I can tell, life happens right at the intersection of two boundaries: the boundary between Newtonian physics and Quantum physics, and the boundary between order and chaos. And the kind of weak, reversible binding between molecules and proteins that you get there is what makes life possible.
But first, let's get a better defintion of what a ligand is...
Ligands
1. The biological function of a protein typically depends on the structure of specific binding sites. These sites are located at the surface of the protein and are determined by geometrical arrangements and physico-chemical properties of tens of non-hydrogen atoms. ...
The ability of proteins to form specific stable complexes is fundamental to biological existence. The interaction between ligand and protein takes place at the surface of the protein. This surface is very complex and convoluted. Furthermore, bound ligands vary greatly in size and properties. The smallest ligands such as O2 and NO consist of two covalently linked atoms showing no or only partial atomic charges. Interactions between proteins and ligands of this type are defined by special arrangement of the electron systems of each participant. Likewise, small ions, e.g. calcium, sodium etc., form a complex compound or similar structure with a few special charged atoms of the protein. A large number of known protein ligands are prosthetic groups, substrates and coenzymes. Their molecular masses lie between 100 and about 2000 Da and their binding sites are larger and more complex. The other end of the size scale is defined by the largest interacting partners of proteins: other biological polymers like nucleic acids, other proteins, and polysaccharides. They show molecular masses from 5000 up to 100,000 Da and more. ...
[The] binding sites of smaller ligands seem to be little caves (grooves, pockets, cavities, depressions) at the surface of proteins. (ref.) 2. A ligand refers to a specific molecule that can bind to a protein. With respect to viruses, a ligand is a protein on the outer coat of a virus that can bind to a receptor protein on the surface of a cell that the virus will infect. Ligands on the surface of a virus can only bind with specific receptor proteins. Different cell types contain different receptor proteins.(ref.) 3. A ligand is simply a molecule which interacts with a protein, by specifically binding to the protein. You should not make the mistake of thinking of a ligand as a relatively small molecule, involved in some obscure biochemical pathway. This narrow view is incorrect. For example consider the case of a repressor protein involved in the regulation of a gene. The repressor protein binds specifically to a section of chromosomal DNA in order to prevent that gene being expressed. In this case, DNA is the ligand (see opposite). This example also illustrates another important aspect of protein ligand binding which is that the interaction must be specific. The interior of the cell contains many different potential ligands and the protein must interact only with the appropriate molecule. So a ligand can be a nucleic acid, polysaccharide, lipid or even another protein. Protein ligand binding is involved in many cell functions including hormone receptors, gene regulation, transport across membranes, the immune response and enzymes catalysis. Although you may not think of an enzyme catalysed reaction as protein ligand binding, initially the enzyme (protein) must bind to the substrate (ligand) before any chemical changes occur. An example shown in the diagram at left is a gene regulating protein which binds specifically to DNA preventing transcription. The two chains of DNA on the left of the picture are shaded differently. There are two polypeptide chains shown on the right of the picture. In this case we can see that the protein which binds the DNA ligand is a dimer i.e. composed of two separate polypeptide chains.
Any binding of a ligand to a protein is also reversible. The physical interactions between a protein and ligand are the same as those between the protein and water molecules or hydrogen ions for example but these latter molecules do not bind to the protein at a specific site. The ligand will however bind to the protein at a specific site. This site has the necessary physical characteristics to make ligand binding favourable.
The specificity of a binding site for a particular ligand can vary however depending on the structure of the protein. For example, the protein may be able to bind two different ligands and each of these ligands will then compete for the proteins binding site. Typically, these different ligand molecules will share some structural or physical properties and thus both be able to fit into the binding site on the protein. This is known as competitive binding.
In some cases, there may be more than one binding site on the protein molecule. Two different ligand or two similar ligands may be able to bind to the protein, one at each binding site. This allosteric binding is quite common in biological systems. Haemoglobin binding to oxygen is perhaps the most commonly cited example of this. You may already be familiar with allosteric regulation of enzyme action.
Summary of Protein Ligand Binding
An example shown in the diagram at left is a gene regulating protein which binds specifically to DNA preventing transcription. The two chains of DNA on the left of the picture are shaded differently. There are two polypeptide chains shown on the right of the picture. In this case we can see that the protein which binds the DNA ligand is a dimer i.e. composed of two separate polypeptide chains.
Any binding of a ligand to a protein is also reversible. The physical interactions between a protein and ligand are the same as those between the protein and water molecules or hydrogen ions for example but these latter molecules do not bind to the protein at a specific site. The ligand will however bind to the protein at a specific site. This site has the necessary physical characteristics to make ligand binding favourable.
The specificity of a binding site for a particular ligand can vary however depending on the structure of the protein. For example, the protein may be able to bind two different ligands and each of these ligands will then compete for the proteins binding site. Typically, these different ligand molecules will share some structural or physical properties and thus both be able to fit into the binding site on the protein. This is known as competitive binding.
In some cases, there may be more than one binding site on the protein molecule. Two different ligand or two similar ligands may be able to bind to the protein, one at each binding site. This allosteric binding is quite common in biological systems. Haemoglobin binding to oxygen is perhaps the most commonly cited example of this. You may already be familiar with allosteric regulation of enzyme action.
Summary of Protein Ligand Binding
Although the driving force for ligand binding is often ascribed to the hydrophobic effect, electrostatic interactions also influence the binding process of both charged and nonpolar ligands. Electrostatic steering of charged substrates into enzyme active sites [conserves] electrostatic potential [energy] localized at the active sites and are the primary determinants of the bimolecular association rates. A more subtle effect is "ionic tethering: salt links can act as tethers between structural elements of an enzyme that undergo conformational change upon substrate binding, and thereby regulate or modulate substrate binding. Ionic tethering can provide a control mechanism for substrate binding that is sensitive to the electrostatic properties of the enzyme's surroundings even when the substrate is nonpolar
Ligands
1. The biological function of a protein typically depends on the structure of specific binding sites. These sites are located at the surface of the protein and are determined by geometrical arrangements and physico-chemical properties of tens of non-hydrogen atoms. ...
The ability of proteins to form specific stable complexes is fundamental to biological existence. The interaction between ligand and protein takes place at the surface of the protein. This surface is very complex and convoluted. Furthermore, bound ligands vary greatly in size and properties. The smallest ligands such as O2 and NO consist of two covalently linked atoms showing no or only partial atomic charges. Interactions between proteins and ligands of this type are defined by special arrangement of the electron systems of each participant. Likewise, small ions, e.g. calcium, sodium etc., form a complex compound or similar structure with a few special charged atoms of the protein. A large number of known protein ligands are prosthetic groups, substrates and coenzymes. Their molecular masses lie between 100 and about 2000 Da and their binding sites are larger and more complex. The other end of the size scale is defined by the largest interacting partners of proteins: other biological polymers like nucleic acids, other proteins, and polysaccharides. They show molecular masses from 5000 up to 100,000 Da and more. ...
[The] binding sites of smaller ligands seem to be little caves (grooves, pockets, cavities, depressions) at the surface of proteins. (ref.) 2. A ligand refers to a specific molecule that can bind to a protein. With respect to viruses, a ligand is a protein on the outer coat of a virus that can bind to a receptor protein on the surface of a cell that the virus will infect. Ligands on the surface of a virus can only bind with specific receptor proteins. Different cell types contain different receptor proteins.(ref.) 3. A ligand is simply a molecule which interacts with a protein, by specifically binding to the protein. You should not make the mistake of thinking of a ligand as a relatively small molecule, involved in some obscure biochemical pathway. This narrow view is incorrect. For example consider the case of a repressor protein involved in the regulation of a gene. The repressor protein binds specifically to a section of chromosomal DNA in order to prevent that gene being expressed. In this case, DNA is the ligand (see opposite). This example also illustrates another important aspect of protein ligand binding which is that the interaction must be specific. The interior of the cell contains many different potential ligands and the protein must interact only with the appropriate molecule. So a ligand can be a nucleic acid, polysaccharide, lipid or even another protein. Protein ligand binding is involved in many cell functions including hormone receptors, gene regulation, transport across membranes, the immune response and enzymes catalysis. Although you may not think of an enzyme catalysed reaction as protein ligand binding, initially the enzyme (protein) must bind to the substrate (ligand) before any chemical changes occur.
Although the driving force for ligand binding is often ascribed to the hydrophobic effect, electrostatic interactions also influence the binding process of both charged and nonpolar ligands. Electrostatic steering of charged substrates into enzyme active sites [conserves] electrostatic potential [energy] localized at the active sites and are the primary determinants of the bimolecular association rates. A more subtle effect is "ionic tethering: salt links can act as tethers between structural elements of an enzyme that undergo conformational change upon substrate binding, and thereby regulate or modulate substrate binding. Ionic tethering can provide a control mechanism for substrate binding that is sensitive to the electrostatic properties of the enzyme's surroundings even when the substrate is nonpolar
Enzymes: Come together, right now, over me.
Definition of an enzyme: Any of numerous proteins or conjugated proteins produced by living organisms and functioning as biochemical catalysts. Enzyme names almost all end in "-ase".
Definition of Catalyst: a substance that decreases the required activation energy to increase the reaction rate. Chemical composition of a catalyst is not altered by the reaction and thus a single catalyst molecule can be used over and over again.
Definition of Kinase: Any of various enzymes that catalyze the transfer of a phosphate group from a donor, such as ADP or ATP, to an acceptor. Kinases alter the shape of acceptor proteins and this change in shape changes which binding sites on the protein molecule are exposed and which other proteins are able to bind with the available sites.
Some key concepts:
Allosteric Enzymes
Conformational Change in Protein From Enzymes are life's catalysts:
Tears convey happiness, or sadness, but they do something else as well, and exactly what was accidentally discovered in the early 1900s. A bacteriologist by the name of Alexander Fleming happened to create an inadvertent experiment when a teardrop fell into one of his bacteria cultures. It set the stage for our modern understanding of how cell processes are controlled. Many years later the chemical in tears was isolated and described. It was an enzyme -- now called Fleming’s Lysozyme. Lyse means to break apart, zyme means enzyme. We now know how Flemming’s Lysozyme works. First, like all enzymes, it’s a protein, a huge molecule built from a perfectly ordered assemblage of amino acid building blocks. Its chemical structure fits molecules in the bacterial cell wall like a lock and key, and once the enzyme locks on, the bacterium’s cell wall comes apart. ...
Enzymes often aren’t assembled by your cells until they are needed for a specific job. How does a cell know when turn on a gene for making a particular enzyme? We know something about this turning-on of genes from experiments with bacteria. The selected subject for study: yogurt. Yogurt is really milk, processed by some friendly bacteria. Breaking down the milk is accomplished by specific enzymes the bacteria produce. Bacteria break down milk sugar, lactose, using an enzyme they produced called galactosidase. When there is no lactose around the bacteria don’t bother making galactosidase, but if they encounter molecules of lactose, they start producing the enzyme. On the bacterium’s DNA is the gene for the galactosidase enzyme. The gene has a start signal. Hanging on to it is a protein called a repressor. The repressor’s molecular structure recognizes the lactose molecule. When a lactose molecule bonds on, the repressor releases its grip. RNA polymerase can then slip in and transcribe the galactosidase gene. Soon the bacterial ribosomes are turning out enzymes that will break down the lactose into something the bacterium can use. In the process the bacteria create a tasty treat for us. Biology online's Enzymes and Chemical Reactions:Metabolism consists of synthesis (anabolism) and breakdown (catabolism) of organic molecules required for cell structure and function. Chemical reactions involve: (1) the breaking of chemical bonds in reactant molecules and (2) making of new chemical bonds to form product molecules. Energy is either added or released as heat during chemical reactions. Enzymes are protein catalysts. (A few RNA molecules also possess catalytic activity). In an enzyme-mediated reaction, an enzyme binds to reactants (substrates) to form an enzyme-substrate complex, which breaks down to release products and the enzyme. The region of the enzyme to which the substrate binds is called the active site, the shape of which determines the chemical specificity of the enzyme. Reversibility of a Reaction: Energy released during a reaction determines reversibility of a reaction. Greater the energy released during a reaction, smaller is the probability of product molecules obtaining this energy and undergoing the reverse reaction to reform the reactants. In such a case, ratio of product to reactant concentration will be large and the reaction will tend to be irreversible. In a reversible chemical reaction, rate of forward reaction decreases and rate of reverse reaction increases as the reaction progresses until the two are equal in a state called the chemical equilibrium, at which point there is no further change in the concentration of reactants and products. Cofactors Substances that bind to enzymes to alter their conformations and make them active. Some cofactors are trace elements. In cases where the cofactor is an organic molecule, it is called a coenzyme. Coenzymes are derived from vitamins. Vitamins From the National Science Teachers SciLink website: Vitamins are organic molecules that function in a wide variety of capacities within the body. The most prominent function is as cofactors for enzymatic reactions. The distinguishing feature of the vitamins is that they generally cannot be synthesized by mammalian cells and, therefore, must be supplied in the diet. More...
The Chemical level of Organization
Allosteric Enzymes
Conformational Change in Protein From Enzymes are life's catalysts:
Tears convey happiness, or sadness, but they do something else as well, and exactly what was accidentally discovered in the early 1900s. A bacteriologist by the name of Alexander Fleming happened to create an inadvertent experiment when a teardrop fell into one of his bacteria cultures. It set the stage for our modern understanding of how cell processes are controlled. Many years later the chemical in tears was isolated and described. It was an enzyme -- now called Fleming’s Lysozyme. Lyse means to break apart, zyme means enzyme. We now know how Flemming’s Lysozyme works. First, like all enzymes, it’s a protein, a huge molecule built from a perfectly ordered assemblage of amino acid building blocks. Its chemical structure fits molecules in the bacterial cell wall like a lock and key, and once the enzyme locks on, the bacterium’s cell wall comes apart. ...
Enzymes often aren’t assembled by your cells until they are needed for a specific job. How does a cell know when turn on a gene for making a particular enzyme? We know something about this turning-on of genes from experiments with bacteria. The selected subject for study: yogurt. Yogurt is really milk, processed by some friendly bacteria. Breaking down the milk is accomplished by specific enzymes the bacteria produce. Bacteria break down milk sugar, lactose, using an enzyme they produced called galactosidase. When there is no lactose around the bacteria don’t bother making galactosidase, but if they encounter molecules of lactose, they start producing the enzyme. On the bacterium’s DNA is the gene for the galactosidase enzyme. The gene has a start signal. Hanging on to it is a protein called a repressor. The repressor’s molecular structure recognizes the lactose molecule. When a lactose molecule bonds on, the repressor releases its grip. RNA polymerase can then slip in and transcribe the galactosidase gene. Soon the bacterial ribosomes are turning out enzymes that will break down the lactose into something the bacterium can use. In the process the bacteria create a tasty treat for us. Biology online's Enzymes and Chemical Reactions:Metabolism consists of synthesis (anabolism) and breakdown (catabolism) of organic molecules required for cell structure and function. Chemical reactions involve: (1) the breaking of chemical bonds in reactant molecules and (2) making of new chemical bonds to form product molecules. Energy is either added or released as heat during chemical reactions. Enzymes are protein catalysts. (A few RNA molecules also possess catalytic activity). In an enzyme-mediated reaction, an enzyme binds to reactants (substrates) to form an enzyme-substrate complex, which breaks down to release products and the enzyme. The region of the enzyme to which the substrate binds is called the active site, the shape of which determines the chemical specificity of the enzyme. Reversibility of a Reaction: Energy released during a reaction determines reversibility of a reaction. Greater the energy released during a reaction, smaller is the probability of product molecules obtaining this energy and undergoing the reverse reaction to reform the reactants. In such a case, ratio of product to reactant concentration will be large and the reaction will tend to be irreversible. In a reversible chemical reaction, rate of forward reaction decreases and rate of reverse reaction increases as the reaction progresses until the two are equal in a state called the chemical equilibrium, at which point there is no further change in the concentration of reactants and products. Cofactors Substances that bind to enzymes to alter their conformations and make them active. Some cofactors are trace elements. In cases where the cofactor is an organic molecule, it is called a coenzyme. Coenzymes are derived from vitamins. Vitamins From the National Science Teachers SciLink website: Vitamins are organic molecules that function in a wide variety of capacities within the body. The most prominent function is as cofactors for enzymatic reactions. The distinguishing feature of the vitamins is that they generally cannot be synthesized by mammalian cells and, therefore, must be supplied in the diet. More...
The Chemical level of Organization
Saturday, February 18, 2006
ATP: Power to the people, right on!
"[The] basic constituents of living matter, such as the amino acids and sugars, are essentially the same in all types of life. The study of intermediary metabiolism shows that the basic metabolic processes, in particular those providing energy, and those leading to the synthesis of cell constituents are also shared by all forms of life. [This suggests that] the mechanism of energy production has arisen very early in the evolutionary process, and secondly, that life, in its present forms has arisen only once."
-- Hans A. Krebs, from the conclusion of the Nobel Lecture he gave when awarded the Nobel prize in 1953.
AMP = Adenosine mono phosphate
ADP = Adenosine di phosphate
ATP = Adenosine tri phosphate
From the University of Hamburg:

The ATP molecule
ATP is regarded as a universal source of energy in all kinds of cells. It is produced mainly in the oxidizing of energy-rich (reduced) compounds in the course of the respiratory chain and in photosynthesis. From Enzymes are life's catalysts:: Adenosine triphosphate is the primary molecule to which energy is transferred during the breakdown of fuel molecules, carbohydrates and fats. (A cell cannot use heat energy to perform its functions). ATP is then hydrolyzed to release energy which can be used by the energy requiring processes in cells such as (1) production of force and movement, (2) active transport across membranes and (2) synthesis of organic molecules used in cell structures and functions. ATP is an energy transfer molecule and NOT an energy storage molecule. It transfers energy from fuel molecules to cells in small amounts OK, so energy is being transferred - but what does this really mean? What exactly is being transported around and how does the body take advantage of this? I've found very interesting answers: 1.) When energy (such as in a photon) is pumped into a chemical system, the energy partitions into thermal and electronic components. The thermal component makes the molecules move faster, and the electronic component increases the number of "high-energy" electronic states. Both energy components will foster molecular organization: the faster the molecules vibrate, rotate, and translate, and the more of them that are in electronic states above ground level, the higher is the probability that the molecules will interact and the more work can be done in organizing them. ...
Living organisms store photon energy in chemical form, and then trickle it down molecular chains to the individual molecular bonding sites. ...
The photon energy is stored in the covalent bonds of glucose -- about 6 quanta of photon in one glucose molecules. From this reservoir, energy then flows along various pathways, nursing everything, all organization and all work. The chemical energy chains that nurse macromolecular organization commonly use ATP as their final link. (ref.) 2.) From Trudy Wassenaar: Many different proteins extract energy from the hydrolysis of ATP. All of them appear to couple some sort of motion of the protein with the energy-releasing hydrolysis. The motion of the protein can in turn be harnessed to do necessary things, such as pump chemicals across a cell membrane, move the cell around, synthesize new proteins, and so on. 3.) Remember the Ion Pump description that won Jens C. Skou the 1997 Nobel prize? Well, he shared the 1997 Nobel Prize in Chemistry with Paul D. Boyer, John E. Walker "for their elucidation of the enzymatic mechanism underlying the synthesis of adenosine triphosphate (ATP). Check out Boyer's lecture "Energy, Life and ATP", and Walker's lecture"ATP Synthesis by Rotary Catalysis". Digging further back in time brings you to the 1953 The Nobel Prize in Physiology or Medicine awarded to Hans Krebs for his work on the Citric Acid Cycle, and Fritz Lipmann for his discovery of "co-enzyme A and its importance for intermediary metabolism". Lipmann talks about "energy-rich phosphate bonds" and "transformations of electron transfer potential ... to phosphate bond energy". John Kyrk's put together two very cool animations that show how the Krebs citric acid cycle works and how mitochondria create ATP. 4.) From University of Connecticut: When phosphate groups are joined together, they have a strong tendency to repel each other, because of the high concentration of negative charge in the very polar and usually ionized oxygen atoms. [the O- in the ATP molecular diagram] As a result, molecules with two or three phosphate groups are good energy donors, readily releasing energy along with the transfer of phosphate groups.
4.) From University of Connecticut: When phosphate groups are joined together, they have a strong tendency to repel each other, because of the high concentration of negative charge in the very polar and usually ionized oxygen atoms. [the O- in the ATP molecular diagram] As a result, molecules with two or three phosphate groups are good energy donors, readily releasing energy along with the transfer of phosphate groups.
 It feels like a number of things that have been discussed in these notes are finally starting to come together. Where else have we seen these phosphate groups before? Oh yeah - the nucleotides that make up DNA. It's not hard to imagine the ionized O- atoms associated with all of those phosphate groups kicking the molecules around energetically, making sure that they encounter the other molecules that they need to form bonds and ligands with. And why don't these O- atoms bond with, say Na+ or K+ ions? For the same reason that we don't find too many NaCl crystals in the body - they are dissolved by water.
More images and text from the great webpage at the University of Hamburg:ATP and Other Nucleoside Triphosphates or: Links Rich in Energy
It feels like a number of things that have been discussed in these notes are finally starting to come together. Where else have we seen these phosphate groups before? Oh yeah - the nucleotides that make up DNA. It's not hard to imagine the ionized O- atoms associated with all of those phosphate groups kicking the molecules around energetically, making sure that they encounter the other molecules that they need to form bonds and ligands with. And why don't these O- atoms bond with, say Na+ or K+ ions? For the same reason that we don't find too many NaCl crystals in the body - they are dissolved by water.
More images and text from the great webpage at the University of Hamburg:ATP and Other Nucleoside Triphosphates or: Links Rich in Energy

Besides the adenosine nucleotide phosphates occur also uracil, cytosine and guanine phosphates: UMP, UDP, UTP, CMP, CDP, CTP, GMP, GDP, GTP. The triphosphate nucleosides of the mentioned compounds including ATP are components of RNA. They are integrated into the polymer by cleavage of pyrophosphate ( = PP). The corresponding desoxyribose derivatives (dATP, dGTP, dCTP....) are necessary for DNA synthesis where dTTP is used instead of dUTP. The terminal phosphate residues of all nucleoside di- and triphosphates are equally rich in energy. The energy set free by their hydrolysis is used for biosyntheses. They share the work equally: UTP is needed for the synthesis of polysaccharides, CTP for that of lipids and GTP for the synthesis of proteins and other molecules. These specificities are the results of the different selectivities of the enzymes that control each of these metabolic pathways.

The ATP molecule
ATP is regarded as a universal source of energy in all kinds of cells. It is produced mainly in the oxidizing of energy-rich (reduced) compounds in the course of the respiratory chain and in photosynthesis. From Enzymes are life's catalysts:: Adenosine triphosphate is the primary molecule to which energy is transferred during the breakdown of fuel molecules, carbohydrates and fats. (A cell cannot use heat energy to perform its functions). ATP is then hydrolyzed to release energy which can be used by the energy requiring processes in cells such as (1) production of force and movement, (2) active transport across membranes and (2) synthesis of organic molecules used in cell structures and functions. ATP is an energy transfer molecule and NOT an energy storage molecule. It transfers energy from fuel molecules to cells in small amounts OK, so energy is being transferred - but what does this really mean? What exactly is being transported around and how does the body take advantage of this? I've found very interesting answers: 1.) When energy (such as in a photon) is pumped into a chemical system, the energy partitions into thermal and electronic components. The thermal component makes the molecules move faster, and the electronic component increases the number of "high-energy" electronic states. Both energy components will foster molecular organization: the faster the molecules vibrate, rotate, and translate, and the more of them that are in electronic states above ground level, the higher is the probability that the molecules will interact and the more work can be done in organizing them. ...
Living organisms store photon energy in chemical form, and then trickle it down molecular chains to the individual molecular bonding sites. ...
The photon energy is stored in the covalent bonds of glucose -- about 6 quanta of photon in one glucose molecules. From this reservoir, energy then flows along various pathways, nursing everything, all organization and all work. The chemical energy chains that nurse macromolecular organization commonly use ATP as their final link. (ref.) 2.) From Trudy Wassenaar: Many different proteins extract energy from the hydrolysis of ATP. All of them appear to couple some sort of motion of the protein with the energy-releasing hydrolysis. The motion of the protein can in turn be harnessed to do necessary things, such as pump chemicals across a cell membrane, move the cell around, synthesize new proteins, and so on. 3.) Remember the Ion Pump description that won Jens C. Skou the 1997 Nobel prize? Well, he shared the 1997 Nobel Prize in Chemistry with Paul D. Boyer, John E. Walker "for their elucidation of the enzymatic mechanism underlying the synthesis of adenosine triphosphate (ATP). Check out Boyer's lecture "Energy, Life and ATP", and Walker's lecture"ATP Synthesis by Rotary Catalysis". Digging further back in time brings you to the 1953 The Nobel Prize in Physiology or Medicine awarded to Hans Krebs for his work on the Citric Acid Cycle, and Fritz Lipmann for his discovery of "co-enzyme A and its importance for intermediary metabolism". Lipmann talks about "energy-rich phosphate bonds" and "transformations of electron transfer potential ... to phosphate bond energy". John Kyrk's put together two very cool animations that show how the Krebs citric acid cycle works and how mitochondria create ATP.
Besides the adenosine nucleotide phosphates occur also uracil, cytosine and guanine phosphates: UMP, UDP, UTP, CMP, CDP, CTP, GMP, GDP, GTP. The triphosphate nucleosides of the mentioned compounds including ATP are components of RNA. They are integrated into the polymer by cleavage of pyrophosphate ( = PP). The corresponding desoxyribose derivatives (dATP, dGTP, dCTP....) are necessary for DNA synthesis where dTTP is used instead of dUTP. The terminal phosphate residues of all nucleoside di- and triphosphates are equally rich in energy. The energy set free by their hydrolysis is used for biosyntheses. They share the work equally: UTP is needed for the synthesis of polysaccharides, CTP for that of lipids and GTP for the synthesis of proteins and other molecules. These specificities are the results of the different selectivities of the enzymes that control each of these metabolic pathways.
Subscribe to:
Posts (Atom)